We provide single directional and bi-directional primer walking of DNA constructs.
Bi-directional Primer Walk
For bi-directional primer walking, DNA sequencing will start with two universal primers and is continued using primers derived from the ends of the newly determined DNA sequences until the whole insert/ fragment is fully sequenced until both ends. This provides 2x reads for the inserted DNA. If there are any ambiguities in the sequence, we will design new primers to resolve them.
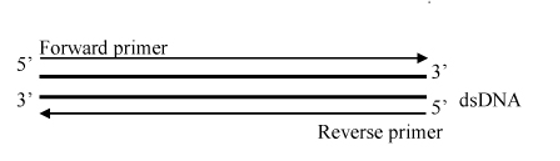
Figure 1: Bi-directional primer walking of construct provides 99.9% accuracy with a double confirmation of the insert sequence on both strands.
Single Pass Primer Walk
For single pass primer walking, DNA sequencing will start with two universal primers and is continued using primers derived from the ends of the newly determined DNA sequences until the overlapping region of both strands are determined. This provides 1x read for the inserted DNA.
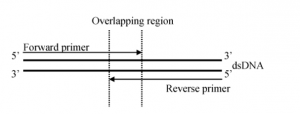
Figure 2: Single pass primer walking of constructs provides a faster turnaround rate with 98.5% accuracy of the insert sequence.
Primer Walking Service Includes:
- Primer design & synthesis
- DNA sequencing reactions using the designed primer(s)
- Data assembly and sequence aligning to generate the consensus sequence
Specifications
The service is charged as per length for the cloned DNA. This service is only recommended for insert sizes less than 7 kb. If you have an inserted DNA > 7 kb, we may process your order as a custom project under our Molecular Biology Services (link to MBS-8001).
During the 1st round of sequencing reactions using the Universal Primers of the holding vector successful, we will send the results to customer to confirm its presence of the inserted DNA. Once customer gives the greenlight to proceed the order, then only we will start the primer walking. We will provide weekly update until the end of the primer walking.
Shall the first round of sequencing reactions failed to meet customer’s expectation, we will terminate the order and only charge for the sequencing reactions.
The base pair charges start from the first until the last base of the inserted fragment in the holding vector. Customer is required to provide vector name and RE cut sites (if this applies) for data alignment.
Contact Us for Orders or Enquiries
Select Your Country
The following is the list of complimentary Universal Primers offered by our DNA Sequencing facility.
| Universal Primers | Sequence (5' - 3') |
| 1492R | 5' TACGGYTACCTTGTTACGACTT 3' |
| 27F | 5' AGAGTTTGATCMTGGCTCAG 3' |
| 35S-A | 5' AAGGGTCTTGCGAAGGATAG 3' |
| 35S-B | 5' AGTGGAAAAGGAAGGTGGCT 3' |
| 518F | 5' CCAGCAGCCGCGGTAATACG 3' |
| 800R | 5' TACCAGGGTATCTAATCC 3' |
| AD Reverse | 5' AGATGGTGCACGATGCACAG 3' |
| a-Factor | 5' TACTATTGCCAGCATTGCTGC 3' |
| AOX1 Forward | 5' GACTGGTTCCAATTGACAAGC 3' |
| AOX1 Reverse | 5' GCAAATGGCATTCTGACATCC 3' |
| BGH-R | 5' TAGAAGGCACAGTCGAGG 3' |
| Bluescript KS | 5' TCGAGGTCGACGGTATC 3' |
| Bluescript SK | 5' CGCTCTAGAACTAGTGGATC 3' |
| CMV-F | 5' CGCAAATGGGCGGTAGGCGTG 3' |
| CMV-profor | 5' ATGGGCGGTAGGCGTG 3' |
| CYC1 Reverse | 5' GCGTGAATGTAAGCGTGAC 3' |
| DsRed1-C | 5' AGCTGGACATCACCTCCCACAACG 3' |
| DsRed1-N | 5' GTACTGGAACTGGGGGGACAG 3' |
| EBV-RP | 5' GTGGTTTGTCCAAACTCATC 3' |
| EGFP-C | 5' CATGGTCCTGCTGGAGTTCGTG 3' |
| EGFP-CF | 5' AGCACCCAGTCCGCCCTGAGC 3' |
| EGFP-CR | 5' CGTCCATGCCGAGAGTG 3' |
| EGFP-N | 5' CGTCGCCGTCCAGCTCGACCAG 3' |
| EGFP-NR | 5' CGTCGCCGTCCAGCTC 3' |
| GAL1 Forward | 5' AATATACCTCTATACTTTAACGTC 3' |
| Gal4AD | 5' TACCACTACAATGGATG 3' |
| GLprimer1 | 5' TGTATCTTATGGTACTGTAACTG 3' |
| GLprimer2 | 5' CTTTATGTTTTTGGCGTCTTCCA 3' |
| HCO2198 | 5' TAAACTTCAGGGTGACCAAAAAATCA 3' |
| hU6-F Primer | 5’-GAGGGCCTATTTCCCATGATT-3’ |
| ITS1 | 5' TCCGTAGGTGAACCTGCGG 3' |
| ITS2 | 5' GCTGCGTTCTTCATCGATGC 3' |
| ITS3 | 5' GCATCGATGAAGAACGCAGC 3' |
| ITS4 | 5' TCCTCCGCTTATTGATATGC 3' |
| ITS5 | 5' GGAAGTAAAAGTCGTAACAAGG 3' |
| KAN2-FP | 5' ACCTACAACAAAGCTCTCATCAACC 3' |
| KAN2-RP | 5' GCAATGTAACATCAGAGATTTTGAG 3' |
| LCO1490 | 5' GGTCAACAAATCATAAAGATATTGG 3' |
| LpJET1.2F | 5' CTGCTTTAACACTTGTGCCTGA 3' |
| LpJET1.2R | 5' TTCCTGATGAGGTGGTTAGCAT 3' |
| M13F (-20) | 5' GTAAAACGACGGCCAGT 3' |
| M13F (-29) | 5' CACGACGTTGTAAAACGAC 3' |
| M13-FP | 5' TGTAAAACGACGGCCAGT 3' |
| M13F-pUC(-40) | 5' GTTTTCCCAGTCACGAC 3' |
| M13R (-20) | 5' GCGGATAACAATTTCACACAGG 3' |
| M13R (-24) | 5' GGAAACAGCTATGACCATG 3' |
| M13R-pUC (-26) | 5' CAGGAAACAGCTATGAC 3' |
| MT Forward | 5' CATCTCAGTGCAACTAAA 3' |
| pBacPAC-RP | 5' GTCTGTAAATCAACAACGC 3' |
| pBAD-F | 5' ATGCCATAGCATTTTTATCCA 3' |
| pBAD-FP | 5' ATGCCATAGCATTTTTATCC 3' |
| pBAD-R | 5' GATTTAATCTGTATCAGG 3' |
| pBRrevBam | 5' GGTGATGTCGGCGATATAGG 3' |
| pDONOR-FP | 5' TAACGCTAGCATGGATCTC 3' |
| pEGFP_N | 5' CCGTCCAGCTCGACCAG 3' |
| pEGFP-FP | 5' TTTAGTGAACCGTCAGATC 3' |
| pEGFP-RP | 5' AACAGCTCCTCGCCCTTG 3' |
| pESP-RP | 5' TCCAAAAGAAGTCGAGTGG 3' |
| pET-24a | 5' GGGTTATGCTAGTTATTGCTCAG 3' |
| pET-RP | 5' CTAGTTATTGCTCAGCGG 3' |
| pFastBac Forward | 5' GGATTATTCATACCGTCCCA 3' |
| pFastBac Reverse | 5' CAAATGTGGTATGGCTGATT 3' |
| pGEX3 | 5' GGAGCTGCATGTGTCAGAGG 3' |
| pGEX5 | 5' GGCAAGCCACGTTTGGTG 3' |
| pJET1.2F | 5' CGACTCACTATAGGGAGAGCGGC 3' |
| pJET1.2R | 5' AAGAACATCGATTTTCCATGGCAG 3' |
| pMalE | 5' TCAGACTGTCGATGAAGC 3' |
| pQE-F | 5' CCCGAAAAGTGCCACCTG 3' |
| pQE-R | 5' GTTCTGAGGTCATTACTGG 3' |
| pREP-fwd | 5' GCTCGATACAATAAACGCC 3' |
| pRH Forward | 5' CTGTCTCTATACTCCCCTATAG 3' |
| pRH Reverse | 5' CAAAATTCAATAGTTACTATCGC 3' |
| pTrcHis Forward | 5' GAGGTATATATTAATGTATCG 3' |
| QE Promoter | 5' CCGAAAAGTGCCACCTG 3' |
| RVprimer3 | 5' CTAGCAAAATAGGCTGTCCC 3' |
| RVprimer4 | 5' GACGATAGTCATGCCCCGCG 3' |
| SP6 | 5' ATTTAGGTGACACTATAG 3' |
| STag 18mer Primer | 5' GAACGCCAGCACATGGAC 3' |
| SV40-pArev | 5' CCTCTACAAATGTGGTATGG 3' |
| SV40-Promoter | 5' GCCCCTAACTCCGCCCATCC 3' |
| T3 | 5' ATTAACCCTCACTAAAG 3' |
| T7 | 5' AATACGACTCACTATAG 3' |
| T7 EEV | 5' ATGTCGTAATAACCCCGCCCCG 3' |
| T7promoter | 5' TAATACGACTCACTATAGGG 3' |
| T7terminator | 5' GCTAGTTATTGCTCAGCGG 3' |
| U-19mer Primer | 5' GTTTTCCCAGTCACGACGT 3' |
| U6 Primer | 5’-GGGCAGGAAGAGGGCCTAT-3’ |
| SP6Long | 5'-ATTTAGGTGACACTATAGAATAC-3' |
| T7Long | 5'-GTAATACGACTCACTATAGGGC-3' |
| LpJet1_2F | 5' CTGCTTTAACACTTGTGCCTGA 3' |
| LpJet1_2R | 5' TTCCTGATGAGGTGGTTAGCAT 3' |
| 785F | 5' GGATTAGATACCCTGGTA 3' |
| 907R | 5' CCGTCAATTCMTTTRAGTTT 3' |
Guides on Preparations and Requirements
![]() Sample Requirements
Sample Requirements![]() List of Services & Details
List of Services & Details![]() Guidelines to Sample Preparation and Quantification
Guidelines to Sample Preparation and Quantification
