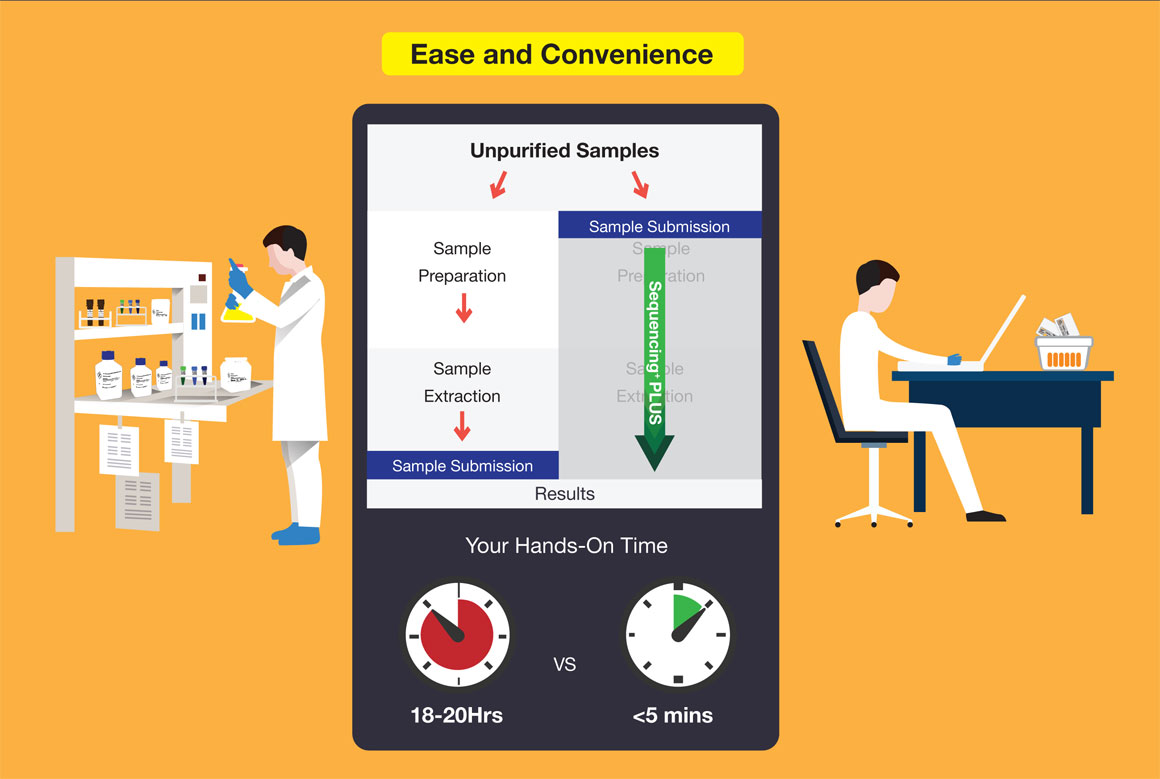
Save the hassle of preparing purified DNA for sequencing and leave your unpurified samples to us. We will do the purification and deliver the best quality results to you.
Whatever sample type you have, we can work with it!
For Unpurified PCR Product
1st BASE Sequencing+ PLUS service allows you to submit PCR products directly off the cycler, saving you the hassle and cost of PCR purification. This service is only recommended for unpurified PCR products that shown single band on agarose gel.
We will perform the PCR clean-up before sequence the purified PCR products directly. Our PCR purification recovery rate is up to 90%. Thus, it is important for you to check the presence of the PCR products before submitting for Sequencing+ PLUS. If additional agarose gel quantification is required before or/ and after the PCR clean-up, a nominal fee will be applied.
For Plasmid
We also accept bacteria culture* for Sequencing+ PLUS service. Send in the cultured broth samples and cell pellet for a fuss free and prompt service, with clean reads >1000bp. Samples which require culturing at our facilities are acceptable as well but might be subjected to a slightly longer turnaround time.
For samples that require culturing at our facilities, we recommend you submit your order latest by Wednesday evening of each week.
*Only vector host by E Coli with Amp/Kanamycin resistance are accepted at our facilities for bacteria culture submission.
*** Applicable for Singapore Customers only.
Contact Us for Orders or Enquiries
Select Your Country
The following is the list of complimentary Universal Primers offered by our DNA Sequencing facility.
| Universal Primers | Sequence (5' - 3') |
| 1492R | 5' TACGGYTACCTTGTTACGACTT 3' |
| 27F | 5' AGAGTTTGATCMTGGCTCAG 3' |
| 35S-A | 5' AAGGGTCTTGCGAAGGATAG 3' |
| 35S-B | 5' AGTGGAAAAGGAAGGTGGCT 3' |
| 518F | 5' CCAGCAGCCGCGGTAATACG 3' |
| 800R | 5' TACCAGGGTATCTAATCC 3' |
| ACYCDuetUP1 | 5' GGATCTCGACGCTCTCCCT 3' |
| AD Reverse | 5' AGATGGTGCACGATGCACAG 3' |
| a-Factor | 5' TACTATTGCCAGCATTGCTGC 3' |
| AOX1 Forward | 5' GACTGGTTCCAATTGACAAGC 3' |
| AOX1 Reverse | 5' GCAAATGGCATTCTGACATCC 3' |
| BGH-R | 5' TAGAAGGCACAGTCGAGG 3' |
| Bluescript KS | 5' TCGAGGTCGACGGTATC 3' |
| Bluescript SK | 5' CGCTCTAGAACTAGTGGATC 3' |
| CMV-F | 5' CGCAAATGGGCGGTAGGCGTG 3' |
| CMV-profor | 5' ATGGGCGGTAGGCGTG 3' |
| CYC1 Reverse | 5' GCGTGAATGTAAGCGTGAC 3' |
| DsRed1-C | 5' AGCTGGACATCACCTCCCACAACG 3' |
| DsRed1-N | 5' GTACTGGAACTGGGGGGACAG 3' |
| DuetDown1 | 5' GATTATGCGGCCGTGTACAA 3' |
| EBV-RP | 5' GTGGTTTGTCCAAACTCATC 3' |
| EGFP-C | 5' CATGGTCCTGCTGGAGTTCGTG 3' |
| EGFP-CF | 5' AGCACCCAGTCCGCCCTGAGC 3' |
| EGFP-CR | 5' CGTCCATGCCGAGAGTG 3' |
| EGFP-N | 5' CGTCGCCGTCCAGCTCGACCAG 3' |
| EGFP-NR | 5' CGTCGCCGTCCAGCTC 3' |
| GAL1 Forward | 5' AATATACCTCTATACTTTAACGTC 3' |
| Gal4AD | 5' TACCACTACAATGGATG 3' |
| GLprimer1 | 5' TGTATCTTATGGTACTGTAACTG 3' |
| GLprimer2 | 5' CTTTATGTTTTTGGCGTCTTCCA 3' |
| HCO2198 | 5' TAAACTTCAGGGTGACCAAAAAATCA 3' |
| hU6-F Primer | 5’-GAGGGCCTATTTCCCATGATT-3’ |
| ITS1 | 5' TCCGTAGGTGAACCTGCGG 3' |
| ITS2 | 5' GCTGCGTTCTTCATCGATGC 3' |
| ITS3 | 5' GCATCGATGAAGAACGCAGC 3' |
| ITS4 | 5' TCCTCCGCTTATTGATATGC 3' |
| ITS5 | 5' GGAAGTAAAAGTCGTAACAAGG 3' |
| KAN2-FP | 5' ACCTACAACAAAGCTCTCATCAACC 3' |
| KAN2-RP | 5' GCAATGTAACATCAGAGATTTTGAG 3' |
| LCO1490 | 5' GGTCAACAAATCATAAAGATATTGG 3' |
| LpJET1.2F | 5' CTGCTTTAACACTTGTGCCTGA 3' |
| LpJET1.2R | 5' TTCCTGATGAGGTGGTTAGCAT 3' |
| M13F (-20) | 5' GTAAAACGACGGCCAGT 3' |
| M13F (-29) | 5' CACGACGTTGTAAAACGAC 3' |
| M13-FP | 5' TGTAAAACGACGGCCAGT 3' |
| M13F-pUC(-40) | 5' GTTTTCCCAGTCACGAC 3' |
| M13R (-20) | 5' GCGGATAACAATTTCACACAGG 3' |
| M13R (-24) | 5' GGAAACAGCTATGACCATG 3' |
| M13R-pUC (-26) | 5' CAGGAAACAGCTATGAC 3' |
| MT Forward | 5' CATCTCAGTGCAACTAAA 3' |
| pBABE 3 | 5' ACCCTAACTGACACACATTCC 3' |
| pBABE 5 | 5' CTTTATCCAGCCCTCAC 3' |
| pBacPAC-RP | 5' GTCTGTAAATCAACAACGC 3' |
| pBAD-F | 5' ATGCCATAGCATTTTTATCCA 3' |
| pBAD-FP | 5' ATGCCATAGCATTTTTATCC 3' |
| pBAD-R | 5' GATTTAATCTGTATCAGG 3' |
| pBRrevBam | 5' GGTGATGTCGGCGATATAGG 3' |
| pDONOR-FP | 5' TAACGCTAGCATGGATCTC 3' |
| pEGFP_N | 5' CCGTCCAGCTCGACCAG 3' |
| pEGFP-FP | 5' TTTAGTGAACCGTCAGATC 3' |
| pEGFP-RP | 5' AACAGCTCCTCGCCCTTG 3' |
| pESP-RP | 5' TCCAAAAGAAGTCGAGTGG 3' |
| pET-24a | 5' GGGTTATGCTAGTTATTGCTCAG 3' |
| pET-RP | 5' CTAGTTATTGCTCAGCGG 3' |
| pFastBac Forward | 5' GGATTATTCATACCGTCCCA 3' |
| pFastBac Reverse | 5' CAAATGTGGTATGGCTGATT 3' |
| pGEX3 | 5' GGAGCTGCATGTGTCAGAGG 3' |
| pGEX5 | 5' GGCAAGCCACGTTTGGTG 3' |
| pJET1.2F | 5' CGACTCACTATAGGGAGAGCGGC 3' |
| pJET1.2R | 5' AAGAACATCGATTTTCCATGGCAG 3' |
| pMalE | 5' TCAGACTGTCGATGAAGC 3' |
| pQE-F | 5' CCCGAAAAGTGCCACCTG 3' |
| pQE-R | 5' GTTCTGAGGTCATTACTGG 3' |
| pREP-fwd | 5' GCTCGATACAATAAACGCC 3' |
| pRH Forward | 5' CTGTCTCTATACTCCCCTATAG 3' |
| pRH Reverse | 5' CAAAATTCAATAGTTACTATCGC 3' |
| pTrcHis Forward | 5' GAGGTATATATTAATGTATCG 3' |
| QE Promoter | 5' CCGAAAAGTGCCACCTG 3' |
| RVprimer3 | 5' CTAGCAAAATAGGCTGTCCC 3' |
| RVprimer4 | 5' GACGATAGTCATGCCCCGCG 3' |
| SP6 | 5' ATTTAGGTGACACTATAG 3' |
| STag 18mer Primer | 5' GAACGCCAGCACATGGAC 3' |
| SV40-pArev | 5' CCTCTACAAATGTGGTATGG 3' |
| SV40-Promoter | 5' GCCCCTAACTCCGCCCATCC 3' |
| T3 | 5' ATTAACCCTCACTAAAG 3' |
| T7 | 5' AATACGACTCACTATAG 3' |
| T7 EEV | 5' ATGTCGTAATAACCCCGCCCCG 3' |
| T7promoter | 5' TAATACGACTCACTATAGGG 3' |
| T7terminator | 5' GCTAGTTATTGCTCAGCGG 3' |
| U-19mer Primer | 5' GTTTTCCCAGTCACGACGT 3' |
| U6 Primer | 5’-GGGCAGGAAGAGGGCCTAT-3’ |
| SP6Long | 5'-ATTTAGGTGACACTATAGAATAC-3' |
| T7Long | 5'-GTAATACGACTCACTATAGGGC-3' |
| LpJet1_2F | 5' CTGCTTTAACACTTGTGCCTGA 3' |
| LpJet1_2R | 5' TTCCTGATGAGGTGGTTAGCAT 3' |
| 785F | 5' GGATTAGATACCCTGGTA 3' |
| 907R | 5' CCGTCAATTCMTTTRAGTTT 3' |
Guides on Preparations and Requirements
![]() Sample Requirements
Sample Requirements![]() List of Services & Details
List of Services & Details![]() Guidelines to Sample Preparation and Quantification
Guidelines to Sample Preparation and Quantification
