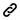The general workflow of NGS include sample preparation, original sample Quality Control (QC), library construction, library Quality Control (QC), sequencing run and bioinformatics analysis. 1st BASE offers NGS services in several categories to meet different needs of customers.
They are: -
Standard NGS Services with/ without Bioinformatics Analysis
Customer provides purified nucleic acid that meets our sample requirements. 1st BASE performs original sample quality control (QC), library construction, library QC, sequencing run with or without bioinformatics analysis.
| Name of Service | Product No. |
| Whole Genome Sequencing (WGS) *For human, animal, plant, fungi, bacterium, virus or insect | NGS-4000 Series |
| Whole Exome Sequencing *For human or mouse | NGS-5000 Series |
| Transcriptome Sequencing *For mRNA, small RNA, lnRNA or Iso-Seq | NGS-6000 Series |
| Metagenomic Sequencing *For environmental samples to target shotgun metagenome, virome, meta-transcriptomic, meta-barcoding, eDNA or any specific region of 16s/ ITS/ 18s gene in amplicon sequencing | NGS-7000 Series |
| Epigenome Sequencing *For Human Whole Genome Bisulfite Sequencing (WGBS) or ChIP (Chromatin Immuno Precipitation) Sequencing | NGS-8000 Series |
| Other Applications *Not limited to but includes custom target capture sequencing, RAD sequencing, organelle sequencing (mtDNA, chloroplast, plasmid), deep amplicon sequencing, fosmid sequencing or BAC sequencing. | NGS-9000 Series |
Sample Preparation and Quantification Service
Customer provides raw materials that meets our sample submission guidelines. 1st BASE performs nucleic acid extraction, purification, sample quality control (QC) with or without library construction services.
** Note: For raw materials not within our submission guidelines, please download Order Form: Sample Preparation Services for non-standard raw material submissions.
| Name of Service | Service Description | Product No. |
| NGS Grade DNA Extraction | 1st BASE to perform DNA extraction, purification, and DNA sample QC. The DNA amount for each order may varies according to different library construction or nature of the project. | NGS-1001 |
| NGS Grade RNA Extraction | 1st BASE to perform total RNA extraction, purification, and RNA sample QC. The RNA amount or RIN requirements for each order may varies according to different library construction or nature of the project. | NGS-1002 |
| Sample QC | Original Sample QC and report. | NGS-1003 |
| DNA Purification | RNase treatment follow by DNA purification. | NGS-1004 |
| RNA Purification | DNase treatment follow by total RNA purification. | NGS-1005 |
| DNA Extraction from Soil/ Stool/ Sludge Samples | Customer to provide raw sample according to our submission guideline. 1st BASE to perform total DNA extraction and purification. The extracted DNA is only suitable for NGS-7000 series of Amplicon Sequencing. | NGS-1007 |
| Custom Primer Synthesis for Amplicon Sequencing | Customer to send amplicons (<500bp) and provide custom primer sequences. Charge per Primer Set to enable Illumina custom amplicon library preparation. | NGS-1008 |
| Library QC | Library QC by qPCR and DNA Fragment Analysis by Agilent TapeStation/ High Sensitivity DNA Labchip/ any equivalent. | NGS-2001 |
| Library Preparation - Illumina platform (Kit dependent) | Library Preparation and with Library Quantification/ Library QC. | NGS-2002 |
| Library Preparation - PacBio platform | Sequel II CLR/ HiFi 10kb Library Preparation and Library QC. | NGS-2003 |
| Library Pooling | Equal molar pooling of different libraries to run in the single sequencing flow cell. | NGS-2004 |
NGS Custom Run Services
Customer provides self-prepared NGS libraries that meets our sample submission guidelines. 1st BASE performs sequencing run with or without library QC services. The sequencing raw data from Illumina MiSeq runs will be provided in Illumina BaseSpace as default with free of charge. *To sign up for a BaseSpace account and retrieving your NGS data from within, please click on the following link for a step by step guide: https://base-asia.com/downloads/products/NGS/Guidelines-for-downloading-NGS-data-from-Illumina-BaseSpace-v2.pdf. The raw data from other sequencing platforms can be provided in Ali Cloud, Amazon Web Services (AWS), Flash Drive or External Hard Disk with separate charges.
| Name of Service | Service Description | Product No. |
| Illumina Single Read Sequencing |
| NGS-3001 |
| Illumina NovaSeq Sequencing (Per Gb) |
| NGS-3003 |
| Illumina NovaSeq Sequencing (Per Lane) |
| NGS-3004 |
| PacBio Sequel II Sequencing (per Gb) |
| NGS-3005 |
| PacBio Sequel II Sequencing (per Cell) |
| NGS-3006 |
| Illumina MiSeq Sequencing v2 (25 PE) |
| NGS-3007 |
| Illumina MiSeq Sequencing v2 (150 PE) Nano Output |
| NGS-3010 |
| Illumina MiSeq Sequencing v2 (250 PE) Nano Output |
| NGS-3012 |
| Illumina MiSeq Sequencing v3 (75 PE) |
| NGS-3013 |
| Illumina MiSeq Sequencing v3 (300 PE) |
| NGS-3014 |
| Nanopore MinION/ GridION Sequencing |
| NGS-3015 |
| Data Delivery | In Illumina BaseSpace/ FTP Link/ Ali Cloud/ AWS/ Flash Drive/ External Hard Disk | NGS-DATA |
For sample submission guidelines and instructions for packing of samples, please visit the "Downloads" tab.