Regular or Standard Fragment Analysis is used to describe genetic marker analysis experiments which rely on detection of changes in the length of a specific DNA Sequence to indicate the presence of absence of a genetic marker. Genetic markers are usually polymorphic genetic sequences contained in or near an allele of interest, such as microsatellite or RFLP, which allow the chromosomal alleles to be distinguished.
At 1st BASE, we offer Standard Fragment Analysis service using Applied Biosystems® Genetic Analyzer to provide the highest resolution at +/- 0.5 bp. The fluorescently label fragment sizes are interpreted using the automated analysis software.
Features of 1st BASE Standard Fragment Analysis Services:
- Free consultation for the experimental design of your genetic marker screening using Fragment Analysis
- DNA size standards up to 500bp for Dye Set D and Dye Set G5 are complementary in our standard fragment analysis services
- Availability of Spectral Optimization before actual run for high accuracy and consistency from different runs
- Stringent automated processes to include control fragment in each of our load on Genetic Analyzers. Only the run with QC passed control fragment(s) are released to our valued customers.
Applications of Standard Fragment Analysis
- Microsatellite Analysis: Simple Sequence Repeats (SSRs), Short Tandem Repeats (STRs)
- AFLP Analysis (amplified fragment length polymorphisms)
- Genotyping
- PCR amplification of genomic DNA using validated conditions
- SNP analysis (single nucleotide polymorphisms)
- Any analysis of fragment size and allele calling
- Electrophoresis of fluorescent PCR products using Genetic Analyzer
DNA Size Standards and Dye Sets
We are using GS500-ROX (for Dye Set D) and GS500-LIZ (for Dye Set G5) DNA Size Standard in our standard fragment analysis services. They consist of 16 DNA fragments, sized as 35, 50, 75, 100, 139, 150, 160, 200, 250, 300, 340, 350, 400, 450, 490 and 500 bp.
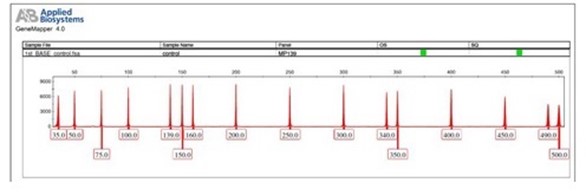
Each band is single-stranded and fluorescently label with ROX/ LIZ, and the size fragments are evenly distributed for very accurate size calling. Your samples will be run together with these known DNA Size Standard for size interpretation using Applied Biosystems® automated analysis software named as Genemapper®.
During the era of manual slab gel electrophoresis system by Applied Biosystems®, the automated fragment analysis software named as GeneScan. Hence, some other sequencing companies will name this as Genescan services.
Dye Set D:
| Dye | Colour |
| FAM | Blue |
| VIC/HEX/JOE/SUN | Green |
| NED/TAMRA | Yellow |
| ROX (Size Standard) | Red |
Dye Set G5:
| Dye | Colour |
| FAM | Blue |
| VIC/HEX/JOE/SUN | Green |
| NED/TAMRA | Yellow |
| PET/ROX | Red |
| LIZ (Size Standard) | Orange |
By having each of your genetic marker labelled with different colour of fluorescent as shown in the tables above, co-loading them together with the known DNA Size Standards; and the Genetic Analyzer will be able to extract individual fluorescent raw data, following by automated analysis software like Genemapper® to interpret the size estimation for each genetic marker. Such high throughout analysis will save your hassle in preparing different gels and collecting the data set by set.
Spectral Optimization
For new set of loci or fluorescently label primer, Spectral Optimization Service is recommended to optimize the loading of samples to obtain optimal signal strength for data analysis. We will include an optimizing step by loading the sample based on separate loci or number of labelled primer(s) at several dilution factors with our DNA Size Standards to determine the optimal amount of sample for subsequent actual run on Genetic Analyzer. The actual run typically consists of co-loading of different fluorescently label primers with different dilution factors (DF). Hence the complexity to get optimal signal strength from each fluorescently label primer in the same run is challenging without spectral optimization.
For customers who multiplex the PCR reactions into a single well or a single tube as multiplexed samples, the level of the spectral optimization will be limited because we are unable to separate the labelled primers to determine the optimum peaks individually.
If spectral optimization is not required, the customer must indicate the Dilution Factor (DF) for each sample in the Order Form. The total volume of ready-to-load sample in our Genetic Analyzer is 10µL. This consist of your PCR sample, our DNA Size Standards and Hi-Di™ Formamide. You will need to know this information before understanding what is meant by the DF in our Order Form.
For example:
DF 10 = if you need us to run 1 µL of your undiluted sample, 1/10 means the DF required is 10.
DF 500 = If the sample needs to be diluted at 50 times before loading, 1µL of the diluted sample to be injected into our Genetic Analyzer will means the DF is 500.
Since all fluorescently label PCR products don’t require purification before loading into Genetic Analyzer for Fragment Analysis, loading more products will introduce more impurities. Instead of expecting higher signal by loading more products, it supresses the collected fluorescent signal during electrophoresis.
To assure all unpurified fluorescently label PCR products doesn’t affect the peak quality of each other in the same capillary run, we only allow maximum of 1uL undiluted sample to be used in Standard Fragment Analysis. This meant, DF 10 will be the maximum dilution factor because we can’t load excess amount of your undiluted (& unpurified) samples into our Genetic Analyzers.
Data Output
Soft copies of .fsa file and .pdf Run Report will be sent through e-mail.
Contact Us for Orders or Enquiries
Select Your Country
Guides on Preparations and Requirements
![]() List of Fragment Analysis Services & Details
List of Fragment Analysis Services & Details![]() Guidelines to Order Preparation and Submission
Guidelines to Order Preparation and Submission
