SNP Genotyping
A single-nucleotide polymorphism (SNP) is a variation in a single nucleotide at a specific location in the genome and is the most frequently occurring genetic variation in living organisms. SNPs are important markers that have been found in numerous studies to have an impact on gene function and phenotype. Hence they have important applications in the medical and agriculture industries.
In humans, most SNPs are normal variants and have no effect on health or development. However, SNP profiling may help in predicting susceptibility to neurological disorders, risk of developing a particular disease or tracking the inheritance of disease genes in families. For example, the presence of two SNPs in the APOE gene codes for 3 different proteins which lead to varying levels of susceptibility for Alzheimer’s disease.
In agriculture, SNPs together with restriction fragment length polymorphisms (RFLP), amplified fragment length polymorphisms (AFLP), simple sequence repeats (SSRs), are used as molecular markers for marker-assisted selection (MAS) in plant breeding. In MAS, individuals or breeds that possess molecular markers which are linked to desired traits (e.g. higher yield, pest or disease resistance) are selected for breeding.
Mutation or Variant Screening
Genome editing is the deliberate alteration of a selected DNA sequence in a living organism by means of insertion, deletion or replacement of the sequence at a selected gene region. This technique incorporates the use of enzymes to cut DNA at targeted locations. Among the recent genome editing technologies, CRISPR-based methods are currently the most promising technique for genetic engineering. There are currently more than 14,000 publications on the CRISPR technology, from less than a hundred in 2011.
The Cas9 enzyme used in the powerful CRISPR approach cuts specific sites with great precision and has the ability to alter a gene at multiple sites in a single procedure, hence making it highly cost-effective as well. Plus, it works on most organisms.
Mutation screening, the detection and characterization of genetic mutations introduced into the host organism, is a major challenge in genome editing as it is tedious and time-consuming. Widely used methods for mutation detection include PCR-based methods such as mismatch cleavage assays and real-time PCR.
As part of the screening process, it is beneficial to first enrich for positive cells transfected with the required plasmids containing a marker such as GFP or antibiotic resistance, followed by a mutation detection assay on the pooled clones population to determine editing efficiency. Sequencing can then be performed on positive clonal cell lines to identify specific clones with the desired edit. This protocol can significantly reduce the overall cost of screening since the researcher is informed whether to proceed to each ensuing step of the experiment.
CRISPR Screening Services
The CRISPR-Cas9 gene editing approach has generated much excitement as a powerful investigative tool in molecular biology and as a driver of precision medicine. Its surging popularity can be attributed to its relative simplicity, superior targeting efficiency and improvements to reduce off-target incidences.
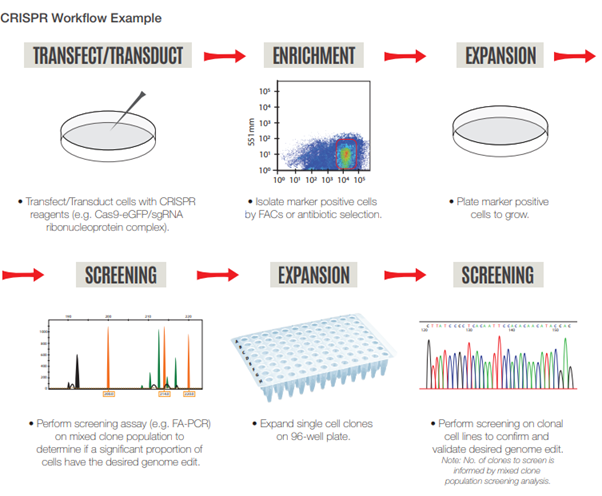
However, researchers have to grapple with challenges in the CRISPR-Cas9 experimental process, particularly in the confirmation of desired on-target mutation events. We offer various methods for INDEL detection for CRISPR screening.
For sample submission guidelines and instructions for packing of samples, please visit the "Downloads" tab.
